Whats behind mysterious cancer hot spots popping up all over the world? (Reuters)
Halfway up a hill overlooking the Great Rift Valley in western Kenya are two graves. One of them is a few years old now, bristling with bushy shrubs stretching bright-green leaves towards a cloudless sky. The other is a freshly dug bed of rough red dirt planted with a white wooden cross. They are the final resting places of Emily’s mother and father, who died within four years of each other.
Still a young woman, Emily now looks after her family’s rural home near Iten — a town famed for churning out long-distance runners and playing host to award-winning British-Somali marathoner Mo Farah’s training camps. We reach it by driving through urban sprawl and out into the hills, passing a seemingly endless stream of impossibly fit athletes pounding the roadside paths.
Emily is busy cooking lunch when we arrive. Her kitchen is a small straw-capped hut built in the traditional style, similar to the other buildings that make up the homestead, with smoke pouring out of the door from an open fire and chickens scratching in the dirt nearby.
It seems idyllic, but there’s a killer on the loose around here: squamous cell oesophageal carcinoma — one of the two main forms of oesophageal cancer.
Cases started piling up more than 60 years ago in South Africa when a doctor working in the then Transkei noticed an unusually high number of people dying from the disease, which was almost unheard of before the 1940s. Since then, scientists have speculated that it might have to do with diet, including the possible contamination of home-grown maize with the rock-bound compound silica from stone mortars. But this has not been proven, a 2017 research review published in the International Journal of Cancer stresses.
Worldwide, an average of 5.9 people per 100 000 will develop oesophageal cancer each year. In Kenya specifically, it’s 18 in 100 000, and in Malawi, it’s even higher — 24 in 100000 — making oesophageal cancer one of the three most common cancers in these countries, a 2017 study in the journal Nature found.
East Africa isn’t the only place in the world where this is happening. The Golestan region of Iran has one of the world’s highest rates of the cancer, and there are pockets of the disease in areas as diverse as Henan province in north-central China and southern Brazil, although it’s relatively rare in neighbouring Colombia, the World Health Organisation’s (WHO’s) Global Cancer Observatory reveals.
Other parts of the world have their own problems: there are strangely high rates of bowel cancer in Slovakia and Denmark, although they have low rates of liver cancer. People in the Czech Republic are more likely to be stricken by kidney or pancreatic cancer than the populations of neighbouring Austria and Poland.
We still don’t really know what’s causing these hot spots. Do these differences lie in inherited genetic variations, in lifestyle or is there an unknown carcinogen lurking in the environment? Maybe it’s a bit of all three?
The wild differences in rates cancer across the world are a mystery — but a crack team of detectives is on the case.
Leading this team is Mike Stratton, director of the Wellcome Sanger Institute near Cambridge, in the United Kingdom. The centre is one of the world’s largest specialising in DNA sequencing and analysis. Together with the International Agency for Research on Cancer (IARC) in Lyon, France — the WHO’s cancer research arm — and others, Stratton has assembled an impressive detective force in cancer research: a project known as Mutographs of Cancer.
By peering deep inside the DNA of cancer cells, Stratton and his team are hunting for the unique signatures or genetic fingerprints that di erent cancer-causing agents and processes leave behind.
The team is recruiting 5 000 people across five continents with five different types of cancer. From them, they’ll extract and analyse DNA from thousands of tumours to build up a massive database of mutational signatures — a bit like Interpol’s international fingerprint collection — so they can try to match causes tcancersrs around the world.
To do this, they will have to create a picture of the genetic mutations that might be associated with cancer. The programme’s name “mutograph” — a combination of “mutation” and “photograph” — is inspired by that mission, the institute’s media officer Samantha Wynne explains.
And their ndings have the potential to save thousands of lives.
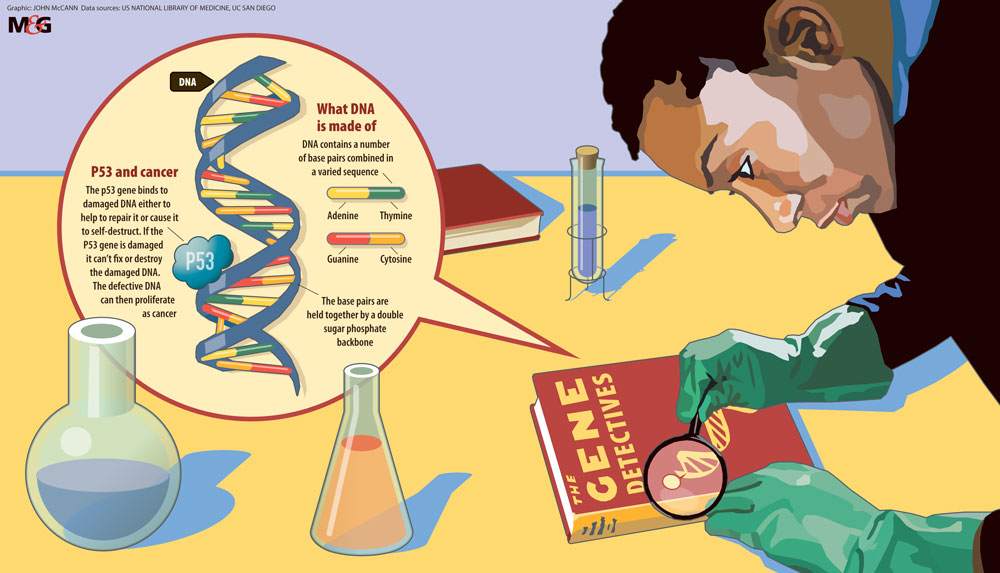
At its heart, cancer is a disease of DNA. The human genome contains 20000 or so genes — the biological instructions that tell our cells when to grow and multiply, what job to do, and even when to die — encoded within long strands of DNA known as chromosomes.
DNA itself is made from four chemical building blocks, or bases, strung together in endlessly varied combinations. It’s the order of these bases — adenine (A), thymine (T), guanine (G) and cytosine (C) — that conveys information within a gene, like a molecular alphabet spelling out the recipes of life. Any changes to the letters in an important gene might cause a cell to start multiplying out of control. Further alterations in other vital genes, along with a cellular environment that allows unchecked growth, will eventually lead to a tumour being formed.
But if you can detect the DNA mutations that give way to cancer and work out what caused them, then you should have the solution to their biological whodunnit.
But to do that, you need to be able to read DNA.
Cancer is a killer and the relationship with it, our genes, our homes and our lives is more than just cause and effect. (John McCann)
In the late 1970s, biochemist Fred Sanger developed a method for reading the sequence of letters in a stretch of DNA. But Sanger’s technique was time-consuming, only allowing scientists to read a few hundred bases at best so researchers focused on just one gene, p53, which is faulty in the majority of human cancers.
By 1991, scientists had shown that different cancers had their own unique suite of mutations in p53, which were likely to have different causes, a study published in the journal Nature found.
Stratton was intrigued.
“Seminal papers suggested that, yes, mutagens that cause cancer left their mark on the genome,” he recalls. “That had a big impression on me as an opportunity. But it’s one that had to be put away in the locker for 15 years waiting for the technology.”
That technology was next-generation sequencing: DNA-reading machines that enable scientists to move from surveying hundreds of bases at a time to thousands or even millions.
Straight away, Stratton saw the potential for the technology to revolutionise our understanding of the genetic changes inside tumours.
By 2009, he and his team had produced the first whole cancer genome sequences. These were detailed maps showing all the genetic changes that had occurred within two individual cancers — a melanoma from the skin and a lung tumour, work that was published a year later in Nature.
These choices of cancer types were far from random: decades of epidemiology and lab studies had shown that UV light exposure is likely to be the strongest sole cause of melanoma, and knowledge of the link between tobacco and lung cancer goes back to the 1950s. With such strong lead suspects, Stratton’s team had the best chance of finding clear mutational fingerprints in the genome. But although they were expecting to see the same kinds of mutations, they weren’t prepared for the scale of genomic vandalism they uncovered.
“The melanoma had something like 25 000 mutations, which was more than the world had ever seen in one genome,” he says.
In the same way that a human fingerprint is a mixture of different patterns of ridges, mutational fingerprints are made up of characteristic patterns of DNA changes.
Carcinogenic chemicals cause mutations by physically binding to specific bases and affecting their shape. These alterations throw a molecular spanner in the works, holding up fundamental processes such as copying DNA, so they have to be fixed to keep the cell healthy and functioning properly. For example, benzo(a)pyrene (one of the major carcinogens in tobacco smoke) tends to bind to G bases, as does aflatoxin, a cancer-causing chemical made by certain moulds. But each of these types of damage is repaired in a specific way, leaving a characteristic change in the DNA sequence.
By contrast, UV light leads to mutations by prompting neighbouring Cs to become stuck together. When the DNA-copying machinery encounters these fused pairs, it interprets the unusual shape as being a pair of Ts, resulting in a permanent change in the DNA sequence in that position.
“In order to analyse and distinguish between these causes, we have to have a way of classifying the patterns of mutations, a bit like identifying a specific set of fingerprints according to the particular patterns of loops and whorls,” Stratton explains.
Initially, Stratton’s team focused on six basic mutational signatures but that wasn’t enough to say whether the changes they found in DNA had caused the cancer. By broadening their scope, they ended up with 96 different subtypes of mutation.
Different mutational processes lead to specific patterns across these 96, which — when graphed — pop out almost as neatly as the ridges and lines of a human fingerprint.
The melanoma and lung cancer genomes were proof that the fingerprints of specific culprits could be seen in cancers with one major cause.
Yet these tumours still contained many mutations that couldn’t be explained by UV or tobacco, so what was causing them? And what about cancers without such an obvious single cause?
With thousands of mutations in a typical tumour, the detective work becomes a lot trickier for cancers with complex, multiple or even completely unknown origins.
By way of analogy, imagine you’re a forensic scientist dusting for fingerprints at a murder scene. You might strike it lucky and find a set of perfect prints on a windowpane that match a known killer in your database.
But you’re much more likely to uncover a mish-mash of fingerprints belonging to a range of people — from the victim and potential suspects to police investigators — all laid on top of each other on all sorts of surfaces.
Fortunately, a former PhD student of Stratton’s, Ludmil Alexandrov, came up with a mathematical method to distinguish mutational fingerprints or signatures from one another, much like splitting out individual vocal and instrumental tracks from a single audio le.
By 2013, Sanger’s team had used a version of this technique to extract 20 distinct signatures from nearly vemillion mutations in more than 7 000 tumours.
Some fingerprints turned up in every single tumour, whereas others were speci c to just a handful of cancers. All of the 30 cancers they looked at had at least two different fingerprints, and some had at least six. That number had risen to around 50 by 2018. A portion of them come from things we already know can increase the risk of cancer, such as tobacco.
Other signatures implicated agents we had previously suspected to cause cancer, but some fingerprints signalled that cancer had been an inside job, a consequence of fundamental processes of life inside our cells.
But the causes of around half these fingerprints remain a mystery, left in the genomes of cancer cells by culprits that are still at large.
What’s behind mysterious cancer hot spots popping up all over the world? (Harriet Lee Merrion, Mosaic)
The endoscopy suite in Moi Teaching and Referral Hospital in Eldoret, western Kenya, is busy. Oesophageal cancer is one of the most common tumours here, and every day a stream of patients arrive in search of relief from the bulging blockages in their gullets. Most have not eaten properly for weeks, making them incredibly thin and frail.
All of them are desperate for help, and most are going to die within the next year.
I watch as little pink globs of cancer tissue are carefully popped into plastic pots and sent o to a freezer, waiting to be shipped to the IARC, then on to the Sanger Institute to be sequenced and analysed for any telltale signatures that might explain what is causing all these cancers.
Diana Menya is a Kenyan epidemiologist at the hospital.
“Some years ago, I noticed that there were quite a number of patients presenting with difficulty swallowing, and when the diagnosis was finally done it was squamous cell oesophageal cancer,” Menya explains. “We were seeing more and more patients.”
Her solution was to set up ESCCAPE (Oesophageal Squamous Cell Carcinoma African Prevention Research), a study that compares the environment and lifestyle of people who have oesophageal cancer to those who don’t.
Menya’s study has already found that tobacco and alcohol are two factors likely to be driving the surfeit of oesophageal cancers — not surprising, given that previous studies have linked them to squamous cell carcinoma.
But these two culprits can’t explain all the cases.
Heading into the rural community outside Iten feels like walking into a world of carcinogens.
For instance, the farmland may be lush and fertile, but it is also awash with pesticides and fertilisers that can leach into the water supply. Sprouting fields of collard greens are cooked into a dish known locally as sukuma wiki (lasts a week) that’s particularly high in nitrates, which the body may convert into carcinogenic nitrosamines when eaten.
We visit Emily in her kitchen; the unventilated room is coated with a thick layer of soot. The smoke from the fire is overpowering.
And where there’s smoke, there’s PAHs — polycyclic aromatic hydrocarbons, a carcinogen released when certain materials such as wood, maize cobs or cow dung burn.
Women, young girls and children are particularly exposed as they spend so much time in the kitchen, often sleeping there at night to keep warm and safe.
Maize is a common food and fuel source in this area and is often treated with fungicide to prevent the growth of toxic pink mould — which may itself cause cancer. Burning these chemicals along with the cobs could release further carcinogens into the unventilated atmosphere.
Although it’s easy to suspect all these things (and more) as being behind the high oesophageal cancer rates in Kenya, we don’t yet have enough data to link most of these risk factors with fingerprints left in the cancer genome. That means we don’t yet know which of them is the most dangerous, or how they might act together to cause disease.
Right now, we’re still in the opening pages of this detective story.
In June 2018, researchers from the Mutographs team shared preliminary data from the first handful of cancer genomes to make it through the project’s pipeline.
Intriguingly, the first few oesophageal tumours from Kenya don’t appear to show any traces of PAHs, potentially putting smoky rural kitchens like Emily’s in the clear, although it’s important to stress that only a small fraction of cases have been analysed so far.
Curiously, all the oesophageal cancers do have signs of damage caused by a family of DNA-altering proteins commonly referred to as APOBECs thought to be activated in response to viral infections.
There’s a lot of scientific hand-waving about the role that these might play in cancer but the discovery of their fingerprints at the biological scene of the crime is intriguing.
Although it’s too early to nail down any suspects for sure, it’s becoming clear from the cancer genomes that we’re not dealing with individual baddies but with a gang of miscreants, each of which administers a potentially fatal blow to the genome.
Each causes mayhem in its own way, but they can combine to bring about catastrophe.
And because every cancer genome is shot through with many thousands of mutations, it’s impossible to say which culprit delivered the coup de grâce. A cell may be riddled with mutations, accumulated from all sorts of processes over a lifetime, but if none of them hit the vital genes or control switches responsible for growth or death, then it will remain healthy.
But we should be able to build up a much better picture of the contributions of different factors — be they biological or environmental — to each individual person’s disease.
This research is coming too late to help Emily’s parents, lying in their hillside graves. But Diana Menya is hopeful that her work will save lives in the future.
“We have eradicated many conditions and especially infectious diseases, so can we not eradicate non-communicable diseases like cancer?” — Additional reporting by Laura López González
This is an edited version of a story originally published by Wellcome on Mosaic.